A protein situated within the flawed a part of a cell can contribute to a number of ailments, reminiscent of Alzheimer’s, cystic fibrosis, and most cancers. However there are about 70,000 completely different proteins and protein variants in a single human cell, and since scientists can usually solely take a look at for a handful in a single experiment, this can be very expensive and time-consuming to determine proteins’ areas manually.
A brand new era of computational strategies seeks to streamline the method utilizing machine-learning fashions that usually leverage datasets containing 1000’s of proteins and their areas, measured throughout a number of cell traces. One of many largest such datasets is the Human Protein Atlas, which catalogs the subcellular habits of over 13,000 proteins in additional than 40 cell traces. However as huge as it’s, the Human Protein Atlas has solely explored about 0.25 % of all attainable pairings of all proteins and cell traces inside the database.
Now, researchers from MIT, Harvard College, and the Broad Institute of MIT and Harvard have developed a brand new computational method that may effectively discover the remaining uncharted area. Their technique can predict the situation of any protein in any human cell line, even when each protein and cell have by no means been examined earlier than.
Their method goes one step additional than many AI-based strategies by localizing a protein on the single-cell degree, relatively than as an averaged estimate throughout all of the cells of a particular sort. This single-cell localization may pinpoint a protein’s location in a particular most cancers cell after remedy, as an example.
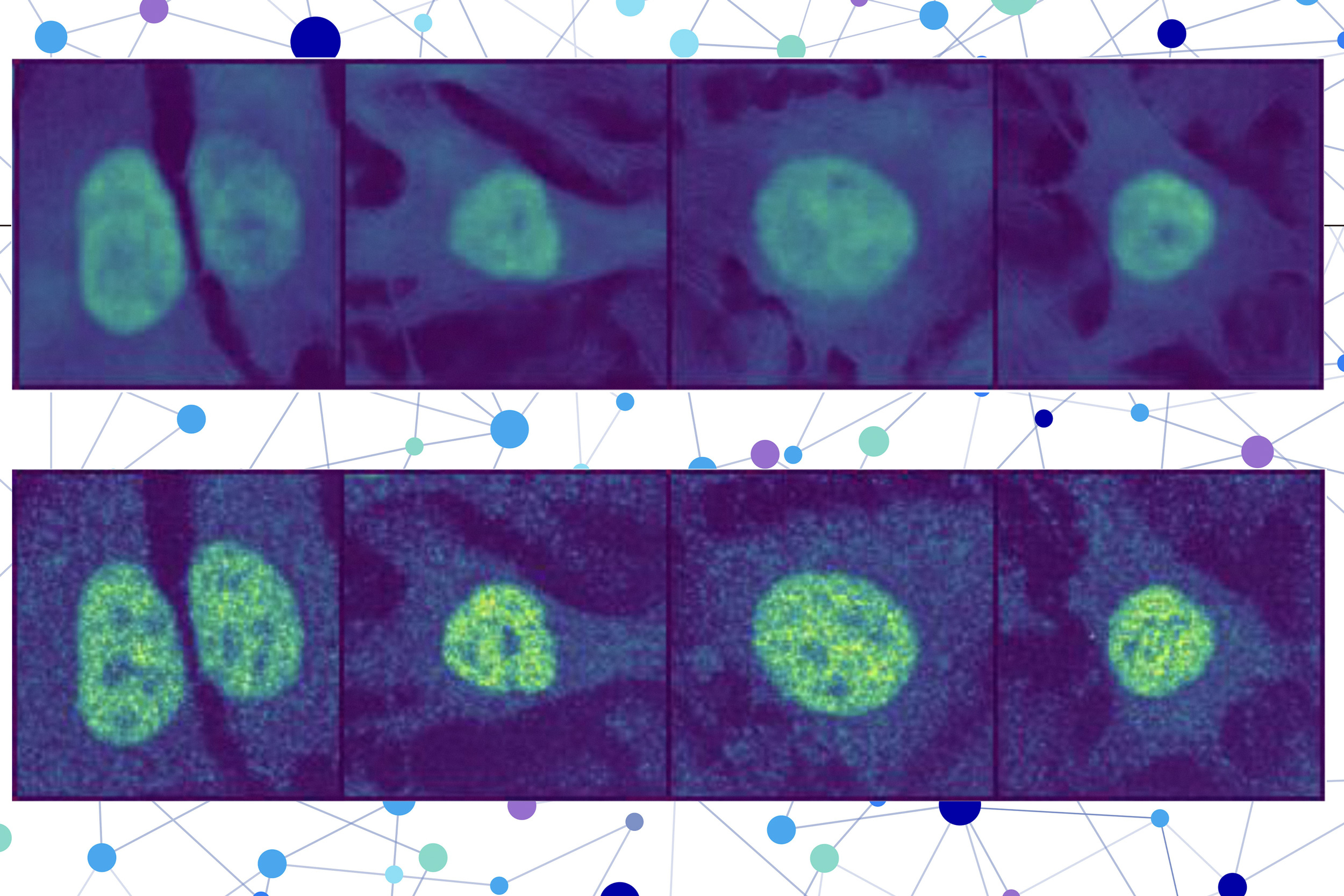
The researchers mixed a protein language mannequin with a particular sort of pc imaginative and prescient mannequin to seize wealthy particulars a couple of protein and cell. Ultimately, the consumer receives a picture of a cell with a highlighted portion indicating the mannequin’s prediction of the place the protein is situated. Since a protein’s localization is indicative of its useful standing, this system may assist researchers and clinicians extra effectively diagnose ailments or determine drug targets, whereas additionally enabling biologists to raised perceive how advanced organic processes are associated to protein localization.
“You would do these protein-localization experiments on a pc with out having to the touch any lab bench, hopefully saving your self months of effort. Whilst you would nonetheless must confirm the prediction, this system may act like an preliminary screening of what to check for experimentally,” says Yitong Tseo, a graduate scholar in MIT’s Computational and Programs Biology program and co-lead creator of a paper on this analysis.
Tseo is joined on the paper by co-lead creator Xinyi Zhang, a graduate scholar within the Division of Electrical Engineering and Laptop Science (EECS) and the Eric and Wendy Schmidt Heart on the Broad Institute; Yunhao Bai of the Broad Institute; and senior authors Fei Chen, an assistant professor at Harvard and a member of the Broad Institute, and Caroline Uhler, the Andrew and Erna Viterbi Professor of Engineering in EECS and the MIT Institute for Information, Programs, and Society (IDSS), who can be director of the Eric and Wendy Schmidt Heart and a researcher at MIT’s Laboratory for Data and Determination Programs (LIDS). The analysis seems at this time in Nature Strategies.
Collaborating fashions
Many current protein prediction fashions can solely make predictions primarily based on the protein and cell information on which they had been skilled or are unable to pinpoint a protein’s location inside a single cell.
To beat these limitations, the researchers created a two-part technique for prediction of unseen proteins’ subcellular location, referred to as PUPS.
The primary half makes use of a protein sequence mannequin to seize the localization-determining properties of a protein and its 3D construction primarily based on the chain of amino acids that varieties it.
The second half incorporates a picture inpainting mannequin, which is designed to fill in lacking components of a picture. This pc imaginative and prescient mannequin appears at three stained photographs of a cell to assemble details about the state of that cell, reminiscent of its sort, particular person options, and whether or not it’s underneath stress.
PUPS joins the representations created by every mannequin to foretell the place the protein is situated inside a single cell, utilizing a picture decoder to output a highlighted picture that exhibits the expected location.
“Totally different cells inside a cell line exhibit completely different traits, and our mannequin is ready to perceive that nuance,” Tseo says.
A consumer inputs the sequence of amino acids that type the protein and three cell stain photographs — one for the nucleus, one for the microtubules, and one for the endoplasmic reticulum. Then PUPS does the remainder.
A deeper understanding
The researchers employed just a few methods through the coaching course of to show PUPS the way to mix data from every mannequin in such a method that it may make an informed guess on the protein’s location, even when it hasn’t seen that protein earlier than.
As an example, they assign the mannequin a secondary job throughout coaching: to explicitly identify the compartment of localization, just like the cell nucleus. That is completed alongside the first inpainting job to assist the mannequin be taught extra successfully.
A great analogy is perhaps a trainer who asks their college students to attract all of the components of a flower along with writing their names. This additional step was discovered to assist the mannequin enhance its normal understanding of the attainable cell compartments.
As well as, the truth that PUPS is skilled on proteins and cell traces on the similar time helps it develop a deeper understanding of the place in a cell picture proteins are likely to localize.
PUPS may even perceive, by itself, how completely different components of a protein’s sequence contribute individually to its general localization.
“Most different strategies often require you to have a stain of the protein first, so that you’ve already seen it in your coaching information. Our method is exclusive in that it may generalize throughout proteins and cell traces on the similar time,” Zhang says.
As a result of PUPS can generalize to unseen proteins, it may seize adjustments in localization pushed by distinctive protein mutations that aren’t included within the Human Protein Atlas.
The researchers verified that PUPS may predict the subcellular location of latest proteins in unseen cell traces by conducting lab experiments and evaluating the outcomes. As well as, when in comparison with a baseline AI technique, PUPS exhibited on common much less prediction error throughout the proteins they examined.
Sooner or later, the researchers wish to improve PUPS so the mannequin can perceive protein-protein interactions and make localization predictions for a number of proteins inside a cell. In the long run, they wish to allow PUPS to make predictions when it comes to residing human tissue, relatively than cultured cells.
This analysis is funded by the Eric and Wendy Schmidt Heart on the Broad Institute, the Nationwide Institutes of Well being, the Nationwide Science Basis, the Burroughs Welcome Fund, the Searle Students Basis, the Harvard Stem Cell Institute, the Merkin Institute, the Workplace of Naval Analysis, and the Division of Power.